Research
The Wang lab is interested in studying how the brain controls behaviors. Our current focus is to understand sleep. Despite being an evolutionarily conserved behavior, sleep remains one of the most intriguing mysteries in biology. We use C. elegans as a model system and exploit its advantages (powerful genetics, optical transparency, and a small nervous system with well-defined anatomical connectivity) to understand the molecular, cellular, and circuit mechanisms underlying sleep. We take an integrative approach by combining classic molecular and genetic tools with state-of-the-art techniques, such as optogenetics, in vivo live imaging, genome editing, and next generation sequencing. In parallel, we also develop novel genetic tools to precisely control transgene expression and manipulate gene activity.
1. Genetic and Circuit Analysis of Sleep
Sleep disorders in humans are prevalent and abnormal sleep can lead to adverse effects on neuronal function and contribute to neurological and other diseases. However, it is unclear how sleep is controlled at the molecular and circuit levels and what sleep does. Our long-term goal is to build a comprehensive picture of the genetic networks and neural mechanisms underlying sleep regulation and function. To understand the conserved biology of sleep, our lab studies a robust sleep behavior in C. elegans that is induced by epidermal growth factor (EGF), a conserved sleep-promoting molecule. EGF signaling induces sleep in C. elegans by acting on a single neuroendocrine neuron. We identified a central neural endocrine mechanism where this neuroendocrine neuron releases multiple neuropeptides to coordinate a sleep state (1). However, it is still unclear what the sleep circuit is, how the sleep circuit is modulated at the molecular level, and how sleep impacts the physiology of the organism. To answer these questions, we will take the advantages of C. elegans: powerful genetics, optical transparency, and a small nervous system with well-defined anatomical connectivity, to systematically study the neural and genetic basis of EGF-induced sleep. Specifically, we are interested in the following three directions:
Sleep disorders in humans are prevalent and abnormal sleep can lead to adverse effects on neuronal function and contribute to neurological and other diseases. However, it is unclear how sleep is controlled at the molecular and circuit levels and what sleep does. Our long-term goal is to build a comprehensive picture of the genetic networks and neural mechanisms underlying sleep regulation and function. To understand the conserved biology of sleep, our lab studies a robust sleep behavior in C. elegans that is induced by epidermal growth factor (EGF), a conserved sleep-promoting molecule. EGF signaling induces sleep in C. elegans by acting on a single neuroendocrine neuron. We identified a central neural endocrine mechanism where this neuroendocrine neuron releases multiple neuropeptides to coordinate a sleep state (1). However, it is still unclear what the sleep circuit is, how the sleep circuit is modulated at the molecular level, and how sleep impacts the physiology of the organism. To answer these questions, we will take the advantages of C. elegans: powerful genetics, optical transparency, and a small nervous system with well-defined anatomical connectivity, to systematically study the neural and genetic basis of EGF-induced sleep. Specifically, we are interested in the following three directions:
- Use brain-wide functional circuit mapping to determine the neural basis of sleep
- Use genetics to identify novel regulators of sleep
- Use transcriptomic analysis and microscopy to understand sleep function
2. Development of Genetic Tools
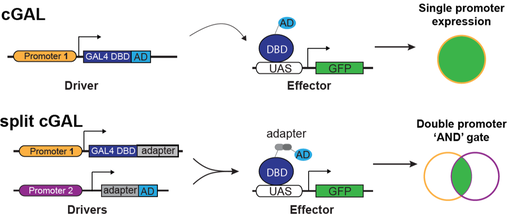
Rigorous neural circuit analyses require powerful genetic tools to precisely control gene expression in specific neurons to manipulate gene and/or neuron activity. A straightforward way to map the sleep circuit in C. elegans is to manipulate each neuron in the entire nervous system and observe the consequences, and/or directly image the activity of each neuron during sleep and awake states. However, our investigation of the neural circuit underlying EGF-induced sleep in C. elegans had been hampered by a lack of such genetic tools that allow efficient and precise transgene control at single neuron resolution. A classic tool is the bipartite GAL4-UAS system, widely used in the Drosophila community. Combining GAL4 drivers and effectors by crossing allows efficient generation of a large number of strains with cell-specific expression of different effectors. We engineered a “cool” GAL4 that works well at low temperatures, and established the first GAL4-based bipartite expression system (cGAL) for C. elegans (2). Since most of the individual C. elegans neurons cannot be genetically targeted using single promoters, we further developed a split cGAL system with an “AND” logic for intersectional targeting using two distinct promoters (3). These tools allow unprecedented genetic access to the C. elegans nervous system at single neuron resolution. The cGAL system can be used to map any other circuits in the worm. Many cGAL driver and effector strains are available (link).
In addition, we also developed a simple and efficient CRISPR/Cas9 genome editing strategy to generate putative null C. elegans mutants by inserting a small universal stop knock-in (STOP-IN) cassette with stop codons in three frames and frameshifts (4). The strategy is cloning-free, with the mixture consisting of preassembled Cas9 ribonucleoprotein and single stranded repair DNA oligos directly injected into gonads of adult C. elegans. The universal STOP-IN cassette also contains a unique sequence that simplifies detection of successful knock-in events via PCR and an exogenous Cas9 target sequence that allows further genome editing. Our lab will continue to expand the genetic toolbox for C. elegans:
Rigorous neural circuit analyses require powerful genetic tools to precisely control gene expression in specific neurons to manipulate gene and/or neuron activity. A straightforward way to map the sleep circuit in C. elegans is to manipulate each neuron in the entire nervous system and observe the consequences, and/or directly image the activity of each neuron during sleep and awake states. However, our investigation of the neural circuit underlying EGF-induced sleep in C. elegans had been hampered by a lack of such genetic tools that allow efficient and precise transgene control at single neuron resolution. A classic tool is the bipartite GAL4-UAS system, widely used in the Drosophila community. Combining GAL4 drivers and effectors by crossing allows efficient generation of a large number of strains with cell-specific expression of different effectors. We engineered a “cool” GAL4 that works well at low temperatures, and established the first GAL4-based bipartite expression system (cGAL) for C. elegans (2). Since most of the individual C. elegans neurons cannot be genetically targeted using single promoters, we further developed a split cGAL system with an “AND” logic for intersectional targeting using two distinct promoters (3). These tools allow unprecedented genetic access to the C. elegans nervous system at single neuron resolution. The cGAL system can be used to map any other circuits in the worm. Many cGAL driver and effector strains are available (link).
In addition, we also developed a simple and efficient CRISPR/Cas9 genome editing strategy to generate putative null C. elegans mutants by inserting a small universal stop knock-in (STOP-IN) cassette with stop codons in three frames and frameshifts (4). The strategy is cloning-free, with the mixture consisting of preassembled Cas9 ribonucleoprotein and single stranded repair DNA oligos directly injected into gonads of adult C. elegans. The universal STOP-IN cassette also contains a unique sequence that simplifies detection of successful knock-in events via PCR and an exogenous Cas9 target sequence that allows further genome editing. Our lab will continue to expand the genetic toolbox for C. elegans:
- Establish a repression cGAL system with a "NOT" gate
- Apply the cGAL system for gene trapping in C. elegans
- Develop genome editing technologies to manipulate gene activity